Redesigned from the ground up, this true next-generation camera is optimized for the most demanding low-dose electron microscopy (EM) applications in both life science and materials science research. The K3® camera series is the complete and latest expression of Gatan’s deep experience in the delivery of real-time, single-electron counting direct detection cameras.
- Powerful inline signal processing will raise the DQE beyond that of the K2® camera; the addition of CDS mode pushes this even higher
- Real-time electron counting immediately lets you know if your samples are good
- Optional inline, GPU-based motion correction avoids the need to save terabytes of raw frames
- 1,500 full frames per second – 3.75 times the speed of the K2 camera
- Match the field of view with your application needs
- K3 (24 megapixels) – Maximize throughput for your highest performance microscopes
- K3 Base (14 megapixels) – Turn screening microscopes into data collection microscopes

-
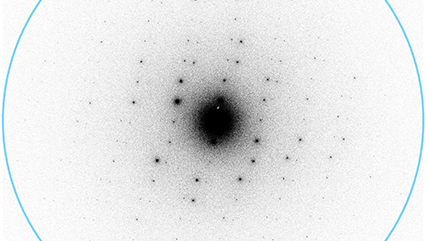
 K3 camera for MicroED/3DED
K3 camera for MicroED/3DED
-
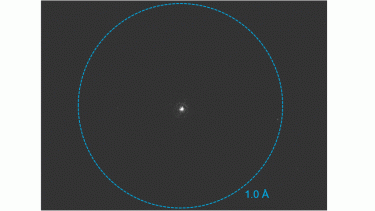
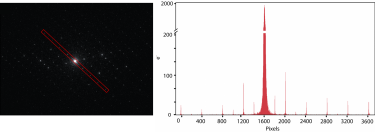 Electron-counted SAED of ZSM-5 with the K3® camera
Electron-counted SAED of ZSM-5 with the K3® camera
-
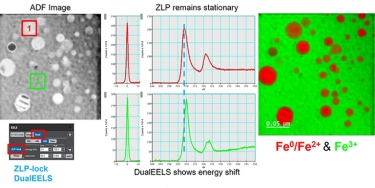
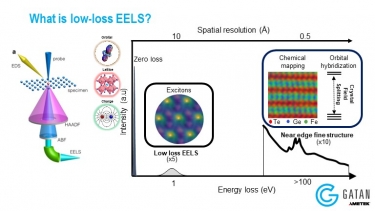
 DualEELS: The importance of low-loss correction of electron energy-loss spectroscopy data
DualEELS: The importance of low-loss correction of electron energy-loss spectroscopy data
-
 Understanding electronic correlations in quantum materials
Understanding electronic correlations in quantum materials
-
 Imaging molecules in their native environment: Cryo-electron tomography of PCDH15 complexes in mouse stereocilia
Imaging molecules in their native environment: Cryo-electron tomography of PCDH15 complexes in mouse stereocilia
-
 Cryo EM reveals mechanisms of gating and drug modulation in 5 HT3A receptors webinar
Cryo EM reveals mechanisms of gating and drug modulation in 5 HT3A receptors webinar
-
 Webinar: High-resolution with the CryoARM/K3 combo: SerialEM, Latitude, and future data collection
Webinar: High-resolution with the CryoARM/K3 combo: SerialEM, Latitude, and future data collection
-

 Using Structural Biology to Drive Pandemic Preparedness Webinar
Using Structural Biology to Drive Pandemic Preparedness Webinar
-

 NUANCE Workshop on 4D STEM: Data Processing using Python
NUANCE Workshop on 4D STEM: Data Processing using Python
-

 NUANCE Workshop on 4D STEM: Data Processing in DM
NUANCE Workshop on 4D STEM: Data Processing in DM
In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges
Turoňová, B.; Sikora, M.; Schürmann, C.; Hagen, W. J. H.; Welsch, S.; Blanc, F. E. C.; von Bülow, S.; Gecht, M.; Bagola, K.; Hörner, C.; van Zandbergen, G.; Landry, J.; de Azevedo, N. T. D.; Mosalaganti, S.; Schwarz, A.; Covino, R.; Mühlebach, M. D.; Hummer, G.; Locker, J. K.; Beck, M.
Molecular architecture of the SARS-CoV-2 virus
Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; Cheng, L,; Shi, D.; Lu, X.; Lei, J.; Crispin, M.; Shi, Y.; Li, L.; Li, S.
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
Wrapp, D.; Wang, N.; Corbett, K. S.; Goldsmith, J. A.; Hsieh, C. -L.; Abiona, O.; Graham, B. S.; McLellan, J. S.
Electron-counting MicroED data with the K2 and K3 direct electron detectors
Clabbers, M. T. B.; Martynawycz, M. W.; Hattne, J.; Nannenga, B. L.; Gonen, T.
Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting
Torino, S.; Dhurandhar, M.; Stroobants, A.; Claessens, R.; Efremov, R. G.
Szemán, A. J. K.; Stráner, P.; Jákli, I.; Hosogi, N.; Harmat, V.; Menyhárd, D. K.; Perczel, A.
Structural basis of actin filament assembly and aging
Oosterheert, W.; Klink, B. U.; Belyy, A.; Pospich, S.; Raunser, S.
Helical ultrastructure of the metalloprotease meprin α in complex with a small molecule inhibitor
Bayly-Jones, C.; Lupton, C. J.; Fritz, C.; Venugopal, H.; Ramsbeck, D.; Wermann, M.; Jäger,C.; Marco, A. D.; Schilling, S.; Schlenzig, D.; Whisstock, J. C.
Stern, A. M.; Yang, Y.; Meunier, A. L.; Liu, W.; Cai, Y.; Ericsson, M.; Liu, L.; Goedert, M.; Scheres, S. H. W.; Selkoe, D.J
Cryo-EM structure of the human NKCC1 transporter reveals mechanisms of ion coupling and specificity
Neumann, C.; Rosenbæk, L. L.; Flygaard, R. K.; Habeck, M.; Karlsen, J. L.; Wang, Y.; Larsen, K. L.; Gad, H. H.; Hartmann, R.; Lyons, J. A; Fenton, R. A.; Nissen, P.
Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex
Naschberger, A.; Mosebach, L.; Tobiasson, V.; Kuhlgert, S.; Scholz, M.; Perez-Boerema, A.; Ho, T. T. H.; Vidal-Meireles, A.; Takahashi, Y.; Hippler, M.; Amunts, A.
Chang, M. R.; Tomasovic, L.; Kuzmina, N. A.; Ronk, A. J.; Byrne, P. O.; Johnson, R.; Storm, N.; Olmedillas, E.; Hou, Y. J.; Schäfer, A.; Leist, S. R.; Tse, L. T.; Ke, H.; Coherd, C.; Nguyen, K.; Kamkaew, M.; Honko, A.; Zhu, Q.; Alter, G.; Saphire, E. O.; McLellan, J. S.; Griffiths, A.; Baric, R. S.; Bukreyev. A.; Marasco. W. A.
Frieg, B.; Geraets, J. A.; Strohäker, T.; Dienemann, C.; Mavroeidi, P.; Jung, B. C.; Kim, W. S.; Lee, S. J.; Xilouri, M.; Zweckstetter. M.; Schröder. G. F.
Lyu, M.; Ayala, J. C.; Chirakos, I.; Su, C. -C.; Shafer, W. M.; Yu, E. W.
Organic crystal growth: Hierarchical self-assembly involving nonclassical and classical steps
Biran, I.; Rosenne, S.; Weissman, H.; Tsarfati, Y.; Houben, L.; Rybtchinski. B.
Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Judge, R. A.; Sridar, J.; Tunyasunvunakool, K.; Jain, R.; Wang, J. C. K.; Ouch, C.; Xu, J.; Mafi, A.; Nile, A. H.; Remarcik, C.; Smith, C. L.; Ghosh, C.; Xu, C.; Stoll, V.; Jumper, J.; Singh, A. H.; Eaton, D.; Hao, Q.
Deagglomeration of DNA nanomedicine carriers using controlled ultrasonication
Hinchliffe, B. A.; Turner, P.; J. H. Cant, D.; De Santis, E.; Aggarwal, P.; Harris, R.; Templeton, D.; Shard, A. G.; Hodnett, M.; Minelli, C.
Structures of a phycobilisome in light-harvesting and photoprotected states
Domínguez-Martín, M. A.; Sauer, P. V.; Kirst, H.; Sutter, M.; Bína, D.; Greber, B. J.; Nogales, E. ;Polívka, T.; Kerfeld, C. A.
Zhu, J.; Huang, W.; Zhao, J., Huynh, L.; Taylor, D. J.; Harris, M. E.
Miyakawa, T.; Yang, J.; Kawasaki, M.; Adachi, N.; Fujii, A.; Miyauchi, Y.; Muramatsu, T.; Moriya, T.; Senda, T.; Tanokura, M.
Identification of mEAK-7 as a human V-ATPase regulator via cryo-EM data mining
Wang, L.; Wu, D.; Robinson, C. V., Fu, T. -M.
Zhao, J.; Makhija, S.; Zhou, C.; Zhang, H.; Wang, Y.; Muralidharan, M.; Huang, B.; Cheng, Y.
Mechanistic details of CRISPR-associated transposon recruitment and integration revealed by cryo-EM
Park, J. U.; Tsai, A. W. -T.; Chen, T. H.; Peters, J. E.; Kellogg, E. H.
Specific recognition and ubiquitination of slow-moving ribosomes by human CCR4-NOT
Absmeier, E.; Chandrasekaran, V.; O'Reilly, F. J.; Stowell, J. A. W.; Rappsilber, J.; Passmore, L. A.
Cryo-electron microscopy of extracellular vesicles
Cai, K.; Sibert, B. S.; Kumar, A.; Yang, J.; Larson, M.; Thompson, K.; Wright, E. R.
Peck, J. V.; Strauss, J. D.; Fay, J. F.
Cryo-EM structure of an active bacterial TIR–STING filament complex
Morehouse, B. R.; Yip, M. C. J.; Keszei, A. F. A.; McNamara-Bordewick, N. K.; Shao, S.; Kranzusch, P. J.
Maintaining the momentum in cryoEM for biological discovery
Thompson, R.; Halfon, Y.; Aspinall, L.; White, J.; Hirst, I. J.; Wang, Y.; Darrow, M.; Muench, S. P.
Structural analysis of the basal state of the Artemis:DNA-PKcs complex
Watanabe, G.; Lieber, M. R.; Williams, D. R.
Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease
Frazier, M. N.; Wilson, I. M.; Krahn, J. M.; Butay, K. J.; Dillard, L. B.; Borgnia, M. J.; Stanley, R. E.
Electron-counting MicroED data with the K2 and K3 direct electron detectors
Clabbers, M. T. B.; Martynowycz, M. W.; Hattne, J.; Nannenga, B. L.; Gonen, T.
Structures and gating mechanisms of human bestrophin anion channels
Owji, A. P.; Wang, J.; Kittredge, A.; Clark, Z.; Zhang, Y.; Hendrickson, W. A.; Yang, T.
Structural insights into dsRNA processing by Drosophila Dicer-2–Loqs-PD
Su, S.; Wang, J.; Deng, T.; Yuan, X.; He, J.; Liu, N.; Li, X.; Huang, Y.; Wang, H. -W.; Ma, J.
Structures and mechanism of the plant PIN-FORMED auxin transporter
Ung, K. L.; Winkler, M.; Schulz, L.; Kolb, M.; Janacek, D. P.; Dedic, E.; Stokes, D. L.; Hammes, U. Z.; Pedersen, B. P.
Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike
Stalls, V.; Lindenberger, J.; Gobeil, S. M. -C.; Henderson, R.; Parks, R.; Barr, M.; Deyton, M.; Martin, M.; Janowska, K.; Huang, X.; May, A.; Speakman, M.; Beaudoin, E.; Kraft, B.; Lu, X.; Edwards, R. J.; Eaton, A.; Montefiori, D. C.; Williams, W. B.; Saunders, K. O.; Wiehe, K.; Haynes, B. F.; Acharya, P.
Structure and flexibility of the yeast NuA4 histone acetyltransferase complex
Zukin, S. A.; Marunde, M. R.; Popova, I. K.; Nogales, E.; Patel, A. B.
Structure and flexibility of the yeast NuA4 histone acetyltransferase complex
Zukin, S. A.; Marunde, M. R.; Popova, I. K.; Nogales, E.; Patel, A. B.
Role of aIF5B in archaeal translation initiation
Kazan, R.; Bourgeois, G.; Lazennec-Schurdevin, C.; Larquet, E.; Mechulam, Y.; Coureux, P. -D.; Schmitt, E.
Ion complexation waves emerge at the curved interfaces of layered minerals
Whittaker, M. L.; Ren, D.; Ophus, C.; Zhang, Y.; Waller, L.; Gilbert, B.; Banfield, J. F.
Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation
Basu, R. S.; Sherman, M. B.; Gagnon, M. G.
Structural Basis for pH-gating of the K+ channel TWIK1 at the selectivity filter
Turney, T. S.; Li, V.; Brohawn, S. G.
Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike
Stalls, V.; Lindenberger, J.; Gobeil, S. M. -C.; Henderson, R.; Parks, R.; Barr, M.; Deyton, M.; Martin, M.; Janowska, K.; Huang, X.; May, A.;l Speakman, M.; Beaudoin, E.; Kraft, B.; Lu, X.; Edwards, R. J.; Eaton, A.; Montefiori, D. C.; Williams, W.; Saunders, K. O.; Wiehe, K.; Haynes, B. F.; Acharya, P.
Structure of S1PR2–heterotrimeric G13 signaling complex
Chen, H.; Chen, K.; Huang, W.; Staudt, L. M.; Cyster, J. G.; Li, X.
Ishii, N.
Structure of the type V-C CRISPR-Cas effector enzyme
Kurihara, N.; Nakagawa, R.; Hirano, H.; Okazaki, S.; Tomita, A.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Scott, D. A.; Nishimasu, H.; Nureki, O.
Zhao, W.; Jensen, G. J.
The giant Mimivirus 1.2 Mb genome is elegantly organized into a 30 nm helical protein shield
Villalta, A.; Schmitt, A.; Estrozi, L. F.; Quemin, E. R. J.; Alempic, J. -M.; Lartigue, A.; Pražák, V.; Belmudes, L.; Vasishtan, D.; Colmant, A. M. G.; Honoré, F. A.; Couté, Y.; Grünewald, K.; Abergel, C.
AAA+ protease-adaptor structures reveal altered conformations and ring specialization
Kim, S.; Fei, X.; Sauer, R. T.; Baker, T. A.
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Zhang, J.; Cai, Y.; Lavine, C. L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M. L.; Rits-Volloch, S.; Wang, S.; Sliz, P.; Wesemann, D. R.; Yang, W.; Seaman, M. S.; Lu, J.; Xiao, T.; Chen, B.
Yu, H.; Hamaguchi, T.; Nakajima, Y.; Kato, K.; Kawakami, K.; Akita, F.; Yonekura, K.; Shen, J. -R.
Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Koenig, P. -D.; Das, H.; Liu H.; Kümmerer, B. M.; Gohr, F. N.; Jenster, L. -M.; Schiffelers, L. D. J.; Tesfamariam, Y. M.; Uchima, M.; Wuerth, J. D.; Gatterdam, K.; Ruetalo, N.; Christensen, M. H.; Fandrey, C. I.; Normann, S.; Tödtmann, J.; M. P.; Pritzl, S.; Hanke, L.; Boos, J.; Yuan, M.; Zhu, X.; Schmid-Burgk, J. L.; Kato, H.; Schindler, M.; Wilson, I. A.; Geyer, M.; Ludwig, K. U.; Hällberg, M.; Wu, N. C.; Schmidt, F. I.
Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants
Wang, L.; Zhou, T.; Zhang, Y.; Yang, E. S.; Schramm, C. A.
Native-like SARS-CoV-2 spike glycoprotein expressed by ChAdOx1 nCoV-19/AZD1222 vaccine
Watanabe, Y.; Mendonça, L.; Allen, E. R.; Howe, A.; Lee, M.; Allen, J. D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; Ulaszewska, M.; Belij-Rammerstorfer, S.; Morris, S.; Krebs, A. -S.; Dejnirattisai, W.; Mongkolsapaya, J.; Supasa, P.; Screaton, G. R.; Green, C. M.; Lambe, T.; Zhang, P.; Gilbert, S. C.; Crispin, M.
Structure of a microtubule-bound axonemal dynein
Walton, T.; Wu, H.; Brown, A.
Mechanism of SARS-CoV-2 polymerase stalling by remdesivir
Kokic, G.; Hillen, H. S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P.
Stabilizing the closed SARS-CoV-2 spike trimer
Juraszek, J.; Rutten, L.; Blokland, S.; Bouchier, P.; Voorzaat, R.; Ritschel, T.; Bakkers, M. J. G.; Renault , L. L. R.; Langedijk, J. P. M.
Zhou, T.; Tsybovsky, Y.; Gorman, J.; Rapp, M.; Cerutti, G.; Chuang, G. -Y.; Katsamba, P. S.; Sampson, J. M.; Schön, A.; Bimela, J.; Boyington, J. C.; Nazzari, A.; Olia, A. S.; Shi, W.; Sastry, M.; Stephens, T.; Stuckey, J.; Teng, I. -T.; Kwong, P. D
Architecture of a SARS-CoV-2 mini replication and transcription complex
Yan, L.; Zhang, Y.; Ge, J.; Zheng, L.; Gao, Y.; Wang, T.; Jia, Z.; Wang, H.; Huang, Y.; Li, M.; Wang, Q.; Ra, Z.; Lou, Z.
An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive spike
Schoof, M.; Faust, B.; Saunders, R. A.; Sangwan, S.; Rezelj, V.; Hoppe, N.; Boone, M.; Billesbølle, C. B.; Puchades, C.; Azumaya, C. M.; Kratochvil, H. T.; Zimanyi, M.; Deshpande, I.; Liang, J.; Dickinson, S.; Nguyen, H. C.; Chio, C. M.; Merz, G. E.; Thompson, M. C.; Diwanji, D.; Schaefer, K.; Anand, A. A.; Dobzinski, N.; Zha, B. S.; Simoneau, C. R.; Leon, K.; White, K. M.; Chio, U. S.; Gupta, M.; Jin, M.; Li, F.; Liu, Y.; Zhang, K.; Bulkley, D.; Sun, M.; Smith, A. M.; Rizo, A. N.; Moss, F.; Brilot, A. F.; Pourmal, S.; Trenker, R.; Pospiech, T.; Gupta, S.; Barsi-Rhyne, B.; Belyy, V.; Barile-Hill, A. W.; Nock, S.; Liu, Y.; Krogan, N. J.; Ralston, C. Y.; Swaney, D. L.; García-Sastre, A.; Ott, M.; Vignuzzi, M.; QCRG Structural Biology Consortium; Walter, P.; Manglik, A.
Zhou, T.; Teng, I. -T.; Olia, A. S.; Cerutti, G.; Gorman, J.; Nazzari, A.; Shi, W.; Tsybovsky, Y.; Wang, L.; Wang, S.; Zhang, B.; Zhang, Y.; Katsamba, P. S.; Petrova, Y.; Banach, B. B.; Fahad, A. S.; Liu, L.; Lopez Acevedo, S. N.; Madan, B.; de Souza, M. O.; Pan, X.; Wang, P.; Wolfe, J. R.; Yin, M.; Ho, D. D.; Phung, E.; DiPiazza, A.; Chang, L. A.; Abiona, O. M.; Corbett, K. S.; DeKosky, B. J.; Graham, B. S.; Mascola, J. R.; Misasi, J.; Ruckwardt, T.; Sullivan, N. J.; Shapiro, L.; Kwong, P. D.
Langer, L. M.; Gat, Y.; Bonneau, F.; Conti, E.
Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2
Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q.
Improved applicability and robustness of fast cryo-electron tomography data acquisition
Eisenstein, F.; Danev, R.; Pilhofer, M.
Surpassing the physical Nyquist limit to produce super-resolution cryo-EM reconstructions
Feathers, J. R.; Spoth, K. A.; Fromme, J. C.
Applications
Models 1025, 1024
Datasheet
Related materials
Nyquist frequency
Dose fractionation and motion correction
Improving DQE with counting and super-resolution
| Modulation transfer function (MTF) curves | ||
|---|---|---|
| 200 kV | 300 kV | |
|
K3 |
CDS | CDS |
| Standard | Standard | |
Related products
BioContinuum® Imaging Filter
Alpine® Direct Detection Camera
K3 IS Camera
Latitude® S Software
Elsa™ Cryo-Transfer Holder
Acknowledgment
Continuing our prosperous collaborations that built the K2, the K3 is the successful result of Peter Denes' team at Lawrence Berkeley National Laboratory and David Agard.