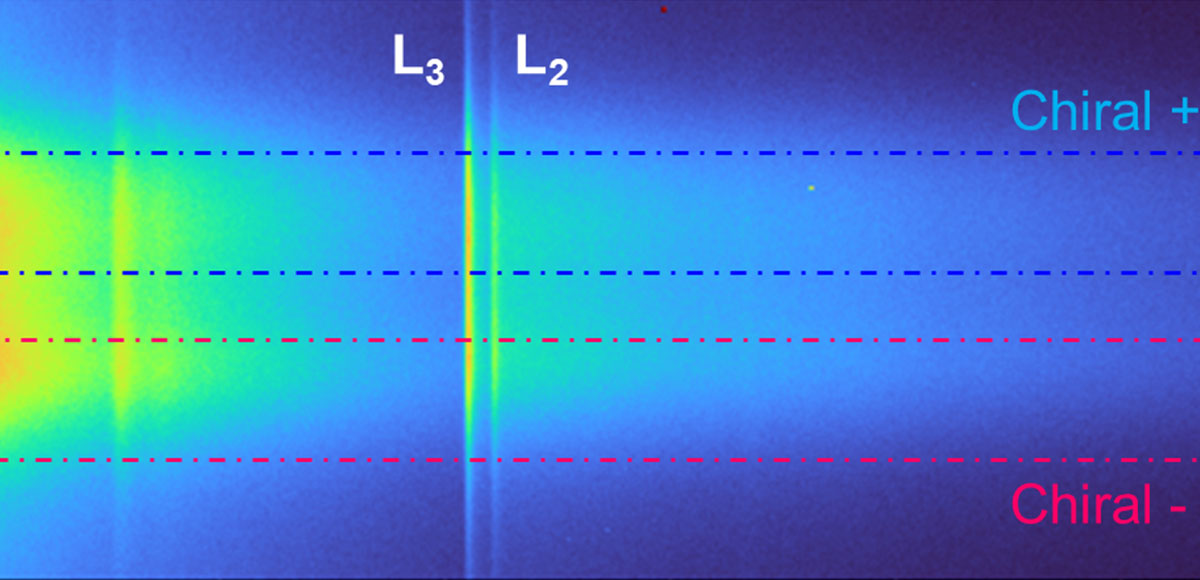
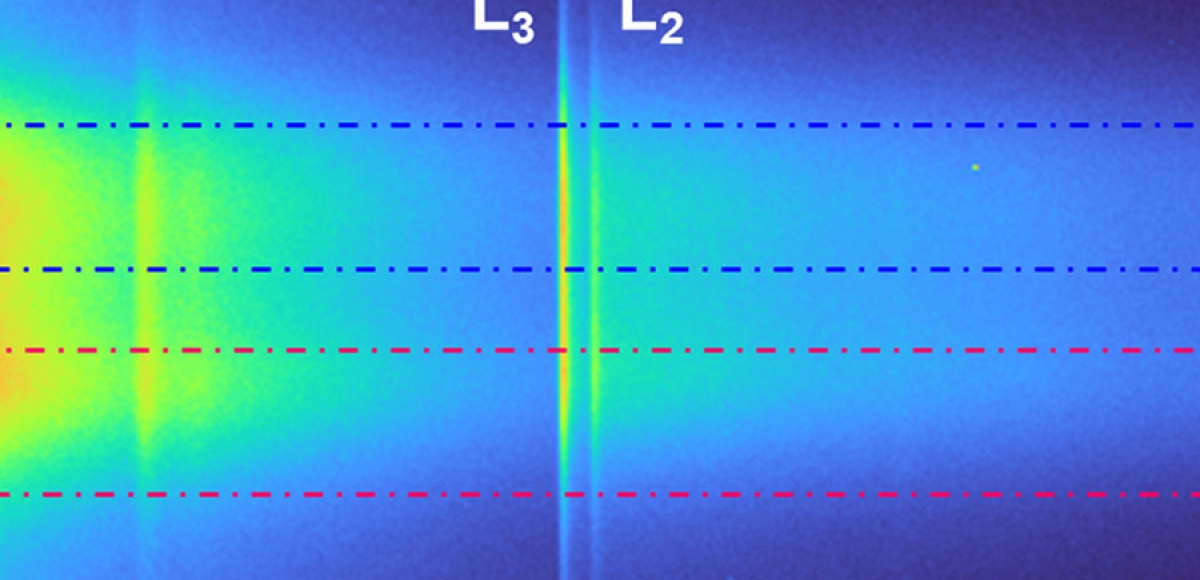
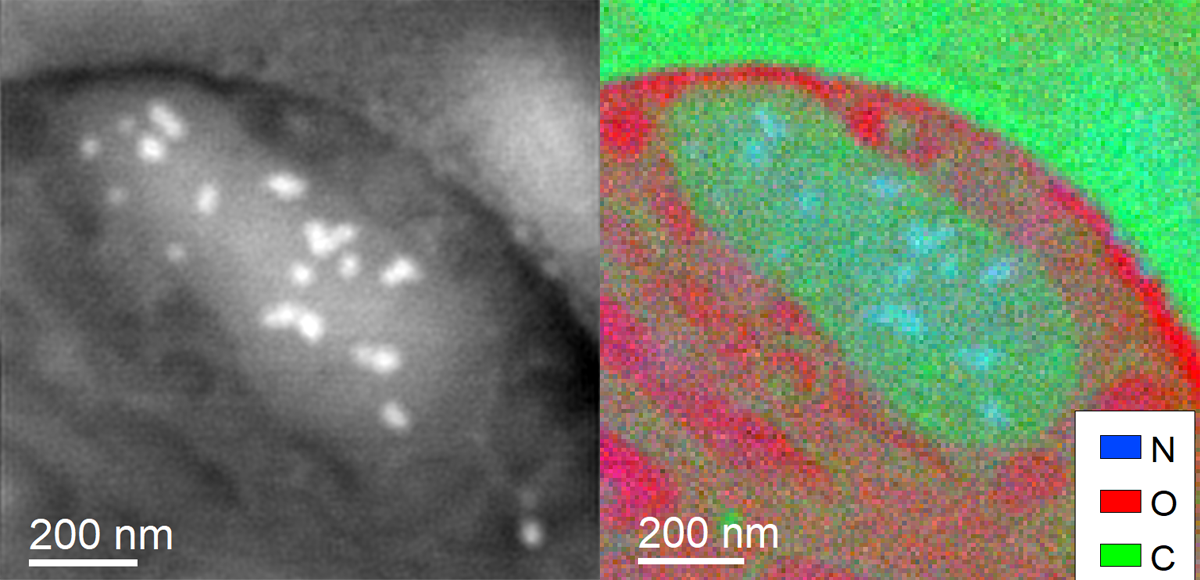
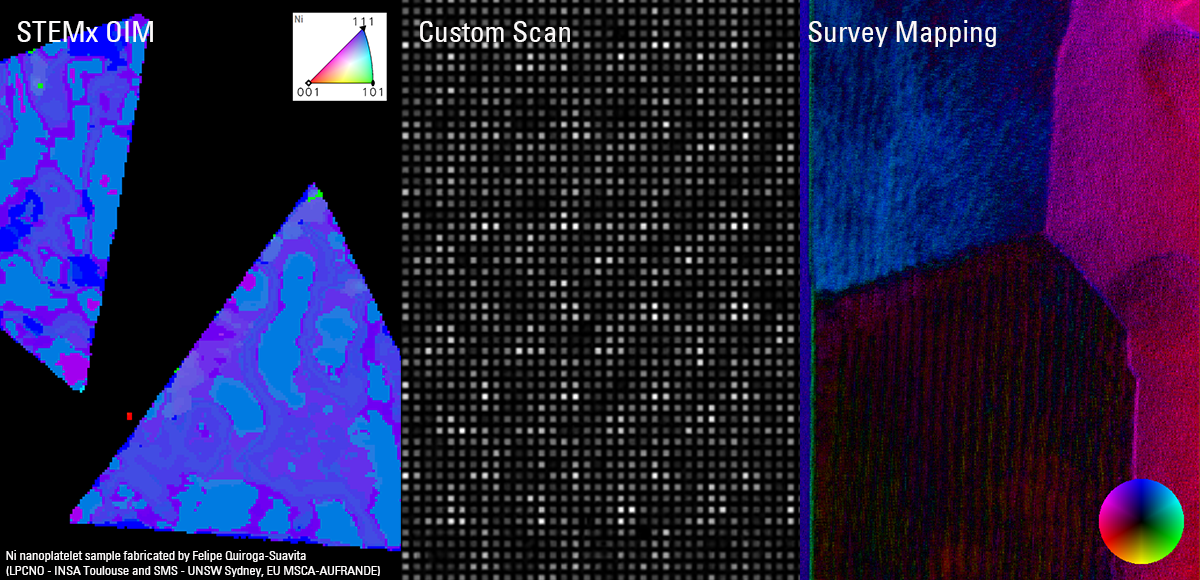
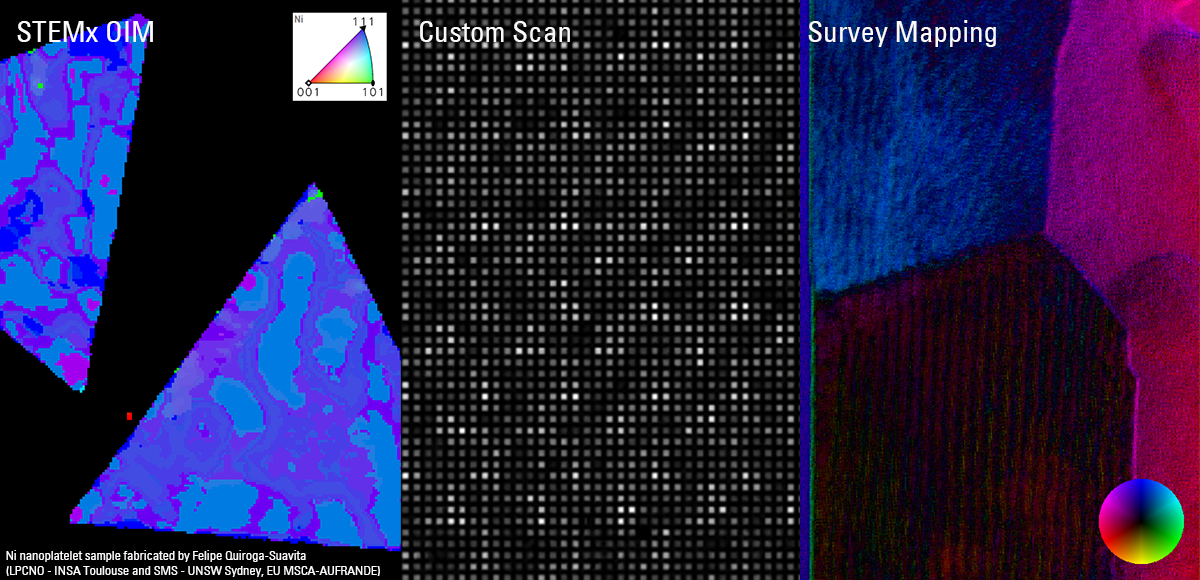
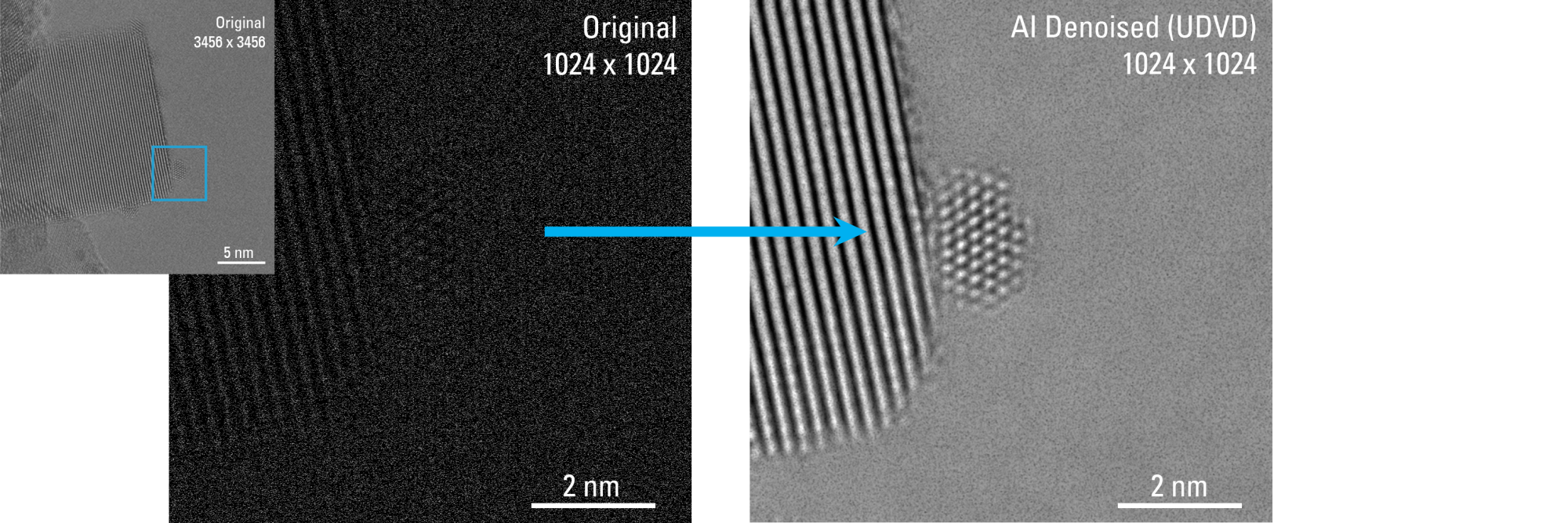
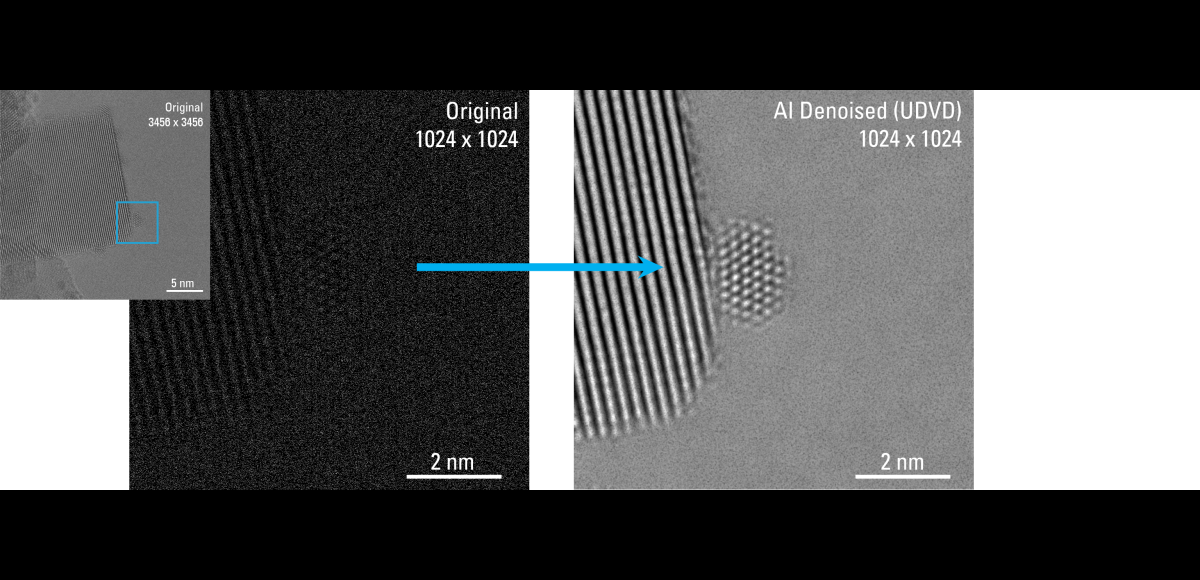
Recent Publications
Phenomenological understanding of the contribution of bulk and grain boundary precipitates on strengthening in prolonged-aged Al-Zn-Mg-Cu aluminium alloys
Materials Today Advances
2025
Strain effects in SrHfO3 films grown by hybrid molecular beam epitaxy
ACS Applied Electronic Materials
2025
NEWS
August 04, 2025
October 29, 2024